The Utrecht Biomolecular Interactions software portal provides access to software tools developed in the Computational Structural Biology group / NMR Research Group of Utrecht University with a main focus on the
characterization of biomolecular interactions. Please note that this site is in active development.
Research
Research within the computational structural biology group focuses on the development of reliable bioinformatic and computational approaches to predict, model and dissect biomolecular interactions at atomic level.
For this, bioinformatic data, structural information and available biochemical or biophysical experimental data are combined to drive the modelling process. By following a holistic approach integrating various experimental information sources with computational structural biology methods we aim at obtaining a comprehensive description of the structural and dynamic landscape of complex biomolecular machines, adding the structural dimension to interaction networks and opening the route to systematic and genome-wide studies of biomolecular interactions.
Services

HADDOCK
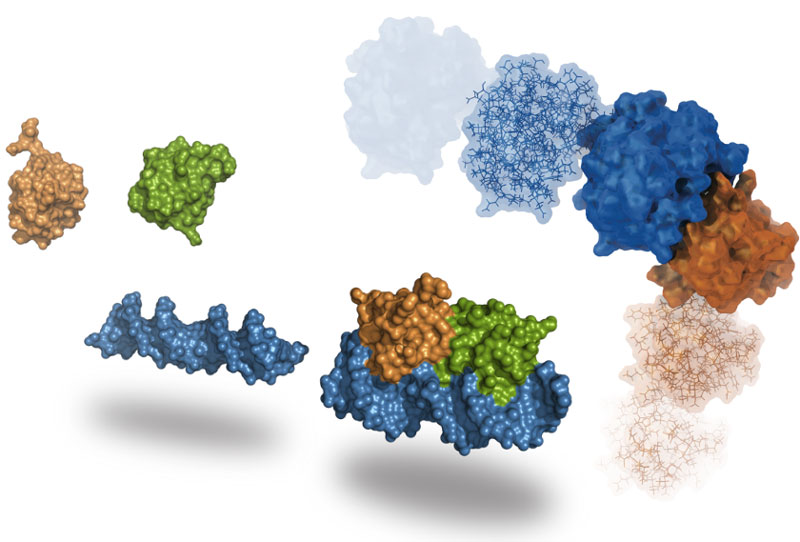
HADDOCK is an integrative platform for the modeling of biomolecular complexes. It supports a large variety of input data and can deal multi-component assembles of proteins, peptide, small molecules and nucleic acids.

PRODIGY
PRODIGY predicts the binding affinity of protein-protein and protein-small molecules complexes and also allows to classify crystallographic interfaces as biological or not.
DISVIS
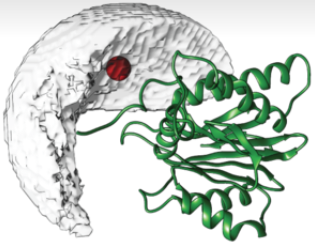
DISVIS quantifies and visualizes the information content of distance restraints between two molecules.
POWERFIT
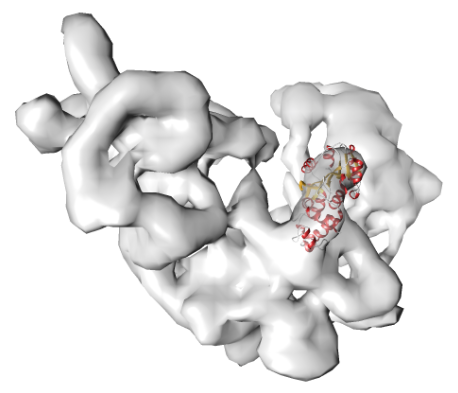
POWERFIT automatically fits atomic models into cryo-EM density maps.

SPOTON
SPOTON determines hot-spot residues at protein-protein interfaces.
CPORT
CPORT is a meta predictor for protein-protein interface residues.

WHISCY
WHISCY predicts protein-protein interfaces, based primarily on conservation, but also taking into account structural information.

3D-DART
3D-DART builds custom DNA models with specific bend properties.

PROABC-2
proABC-2 predicts which residues of an antibody are forming intermolecular contacts with its cognate antigen.
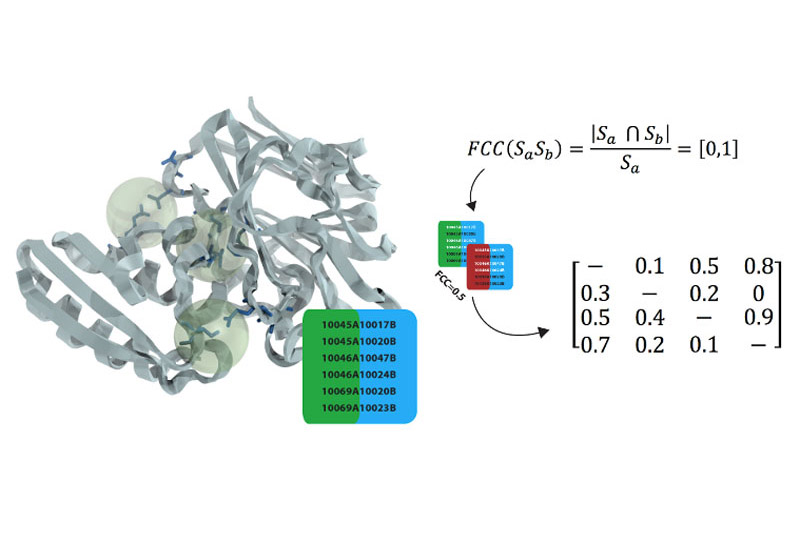
FCC CLUSTERING
A novel clustering strategy that is based on a very efficient similarity measure - the fraction of common contacts.
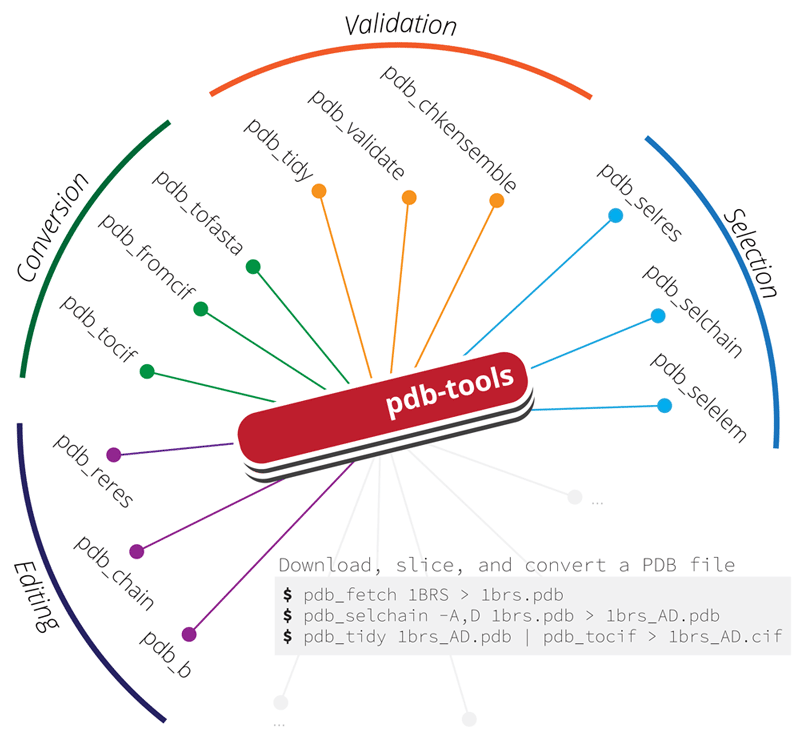
PDB-tools
A dependency-free cross-platform Swiss army knife for massaging PDB files.

GenTBL
Generate HADDOCK compatible ambiguous restraints.

FANDAS
Fast Analysis of multidimensional NMR DAta Sets (FANDAS) is a tool to predict peaks for multidimensional NMR experiments on proteins.
Arctic3D
An algorithm for Data Mining and Clustering of protein interfaces
Benchmarks and Datasets
Protein-DNA Docking Benchmark
Our protein-DNA benchmark contains 47 unbound-unbound test cases of a varying degree of difficulty. Visit the site to read more and download the benchmark.
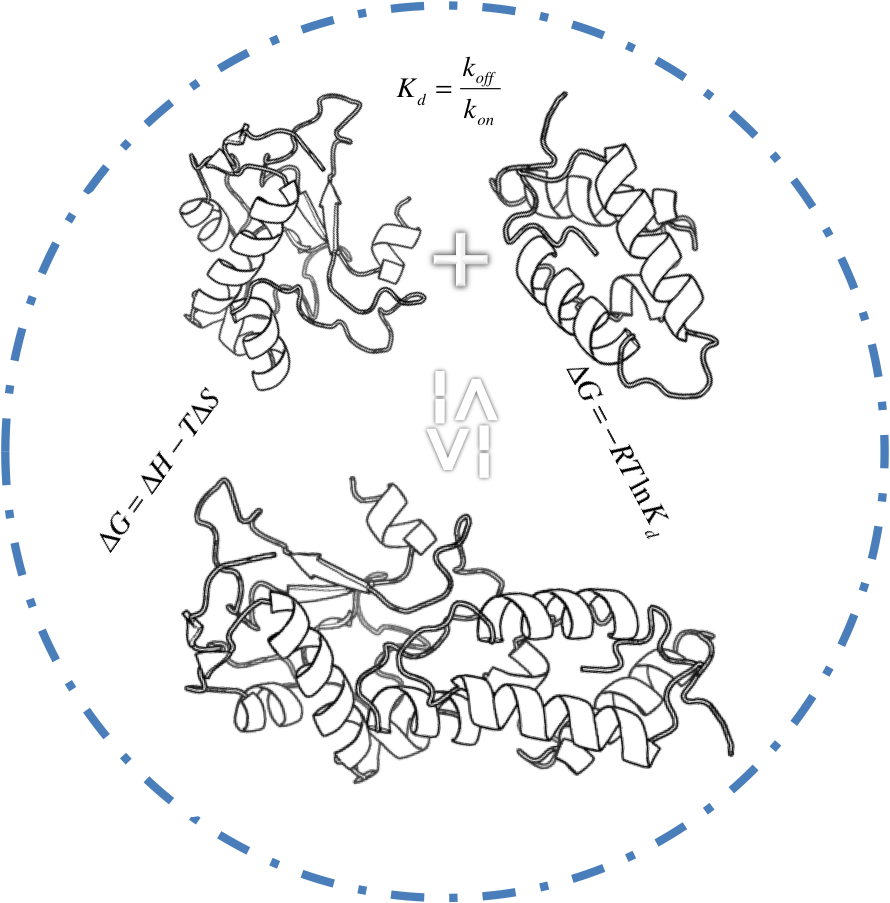
Protein-Protein Binding Affinity Benchmark
We present a protein−protein binding affinity benchmark consisting of binding constants (Kd’s) for 81 complexes.

HADDOCK models
Access various HADDOCK models datasets from our lab page at SBGrid.

HADDOCK-ready BM5
Docking benchmark 5 (BM5) - cleaned and ready to use for HADDOCK.





